In which biochemists observe models of giant molecules as they are displayed on a screen by a computer and try to fold them into the shapes that they assume in nature.
Many problems of modern biology are concerned with the detailed relation between biological function and molecular strocture. Some of the questions currently being asked will be completely answered only when one has an understanding of the structure of all the molecular components of a biological system and a knowledge of how they interact. There are, of course, a large number of problems in biology into which biologists hnve some insight but concerning which they cannot yet ask suitable questions in terms of molecular structure. As they see such problems more clearly, however, they invariably find an increasing need for structural information. In our laboratory at the Massachusetts Institute of Technology we have recently started using a computer to help gain such information about the structure of large biological molecules.
For the first half of this century the metabolic and structural relations among the small molecules of the living cell were the principal concern of biochemists. The chemical reactions these molecules undergo have been studoed intensively. Such reactions are specifically catalyzed by the large proteiu molecules called enzymes, many of which have now been purified and also studied. It is only within the past few years, however, that X-ray-diffraction techniques have made it possible to determine the molecular structure of such protein molecules. These giant molecules, which contain from a thousand to tens of thousands of atoms, constitute more than ha!f of the dry weight of cells. Protein molecules not only act as enzymes but also provide many of the cell's structural components. Another class of giant molecules, the nucleic acids, determine what kind of protein the cell can produce, but most of the physiological behavior of a cell is determined by the properties of its proteins.

The X-ray-diffraction methods for invcstigating the three-dimensional structure of protein molecules are difficult and time-consuming. So far the structures of only three proteins have been worked out: myoglobin. hemoglobin and lysozyme [see "The Three-dimensional Structure of a Protein Molecule," by John C. Kendrew, SCIENTIFIC AMERICAN, December, 1961, and "The Hemoglobin Molecule," by M. F. Perutz, November, 1964]. In their studies of the hemoglobin molecule, M. F. Perutz and his associutes at the Laboratory of Molecular Biology in Cambridge, England, have observed that the structure of the molecule changes slightly when oxygen is attached to it or removed from it. The hemoglobin molecule is the only one for which this kind of study has as yet been carried out. It is known, however, that many proteins change their shape as they perform their functions and that their shape is further modified by the action of the small molecules that activate or inhibit them. The large number of enzyme systems involved in regulating the complex metabolic pathways of the liviug cell have been studied so for only at the level of the overall shape of the enzyme moleule; practically nothing is known of the specific structural changes that may be important for enzyme function and control.





Another problem currently being investigated by many workers concerns the way in which proteins achieve their final three-dimensional configuration when they are synthesized. During the past few years many of the processes involved in protein synthesis have become rather well understood. As a result one knows, at least in general terms, how the cell determines th sequence of amino acids from which protein molecules are assembled and how this sequence establishes the way in which the atoms of a protein are connected [see Figure 2]. It is not, however, the chemical sequence, or connectedness, that establishes the functional properties of a protein. These properties are a consequence of the exact three-dimensional arrangement of the molecule's atoms in space.
As a result of work in the past 15 years, there is now a considerable body of evidence showing that the three-dimensional configuration of a protein molecule is determined uniquely by its amino acid sequence. The number of possible sequences is immense because the cell has at its disposal 20 kinds of amino add building block. The configuration assumed by any particular sequence reflects the fact that the molecule arranges itself so as to minimize its total free energy. In other words, each protein has the shape it has because outside energy would be needed to give it any other shape. The experimental evidence for this conclusion comes from results obtained with many different proteins.
The first really critical experiments in this regard were carried out by Christian B. Anfinsen and his collaborators at the National fInstitutes of Health with the enzyme rlbonuclease, a protein consisting of 153 amino acids in a single chain. In our laboratory we have studied the enzyme alkaline phosphatase, which consists of about 800 amino acids arranged in two chains. Both proteins can be treated so that they lose their well-defined three-dimensional configuration without breaking any of the chemical bonds that establish the connectedness of the molecule. In this "denatured" state the proteins are no longer enzymatically active. But if the denaturing agent is removed and the proteins are put in a solution containing certain salts and the correct acidity, the activity can be reestablished.
The alkaline phosphatase molecule has two identical subunits that are inactive by themselves. They can be separated from each other by increasing the acidity of the solution, and they reassemble to form the active "doublet" when the solution is made neutral once again. In addition the subunits themselves can be denatured, with the result that they become random, or structureless, coils. Under the proper conditions it takes only a few minutes to reestablish the enzymatic activity from this disrupted state, along with what appears to be the original three-dimensional configuration of the doublet molecule. An enzymatically active hybrid molecule can even be formed out of subunits from two different organisms. The individual subunits from the two organisms have different amino acid sequences, but they fold into a shape such that the subunits are still able to recognize each other and form an active molecule. These renaturation processes can take place in a solution containing no protein other than the denatured one, and without the intervention of other cellular components.
Apart from the renaturation experiments, the mechanism of synthesis has suggested an additional factor that may be relevant in establishing the correct three-dimensional form of the protein. It is known that the synthetic process always begins at a particular end of the protein molecule - the end carrying an amino group (NH2) - and proceeds to the end carrying a carboxyl group (COOH). It is plausible to imagine that proteins fold as they are formed in such a way that the configuration of th amino end is sufficiently stable to prevent its alteration while the rest of the molecule is being synthesized. Although this hypothetical mechanism seems to be contradicted by the renaturation experiments just described, it may represent the way some proteins are folded. Because the mechanism would place certain constraints on the folding of a protein molecule, it implies that the active protein is not in a state in which free energy is at a minimum but rather is in a metastable, or temporarily stable, stale of somewhat higher energy.
A molecular biologist's understanding of a molecular structure is usually reflected in his ability to construct a three-dimensional model of it. If the molecule is large, however, model-building can be frustrating for purely mechanical reasons; for example, the model collapses. In any event, the building of models is too time-consuming if one wishes to examine many different configurations, which is the case when one is attempting to predict an unknown structure. When one is dealing with the largest molecules, even a model is not much help in the task of enumerating and evaluating all the small interactions that contribute to the molecule's stability. For this task the help of a computer is indispensable.
Any molecular model is based on the nature of the bonds that hold particular kinds of atoms together. From the viewpoint of the model-builder the important fact is that these bonds are the same no matter where in a large molecule the atom is found. For instance. if a carbon atom has four other atoms bonded to it, they will be arranged as if they were located at the corners of a tetrahedron, so that any two bonds form an angle of approximately 109.5 degrees. The lengths of bonds are even more constant than their angles; bonds that are only one to two angstrom units long are frequently known to be constant in length with an accuracy of a few percent. In general the details of atomic spacing are known from the X-ray analysis of small molecules; this knowledge simplifies the task of building models of large molecules.
An example of the value of such knowledge was the discovery by Linus Pauling and Robert B. Corey at the California Institute of Technology that the fundamental repeating bond in protein structures-the peptide bond that joins the CO of one amino acid to th NII of the next-forms an arrangement of six atoms that lie in a plane. This knowledge enabled them to predict that the amino acid units in a protein chain would tend to become arranged in a particular helical form: the alpha helix. It was subsequently found that such helixes provide a significant portion of the structure of many protein molecules. Thus in advance of any crystallographic information about the structure of a particular protein molecule, one knows the spatial arrangement of atoms within the peptide bonds, as well as the detailed geometry of its alpha-helical regions.
The planar configuration of the peptide bond allows an enormous reduction in the number of variables necessary for a complete description of a protein molecule. Instead of three-dimensional coordinates for each atom, all one needs in order to establish the path of the central chain of the molecule are the two rotation angles where two peptide bonds come together [Figure 8]. The complete description requires, in addition to this information, the specification of the rotation angles of the side chains in those amino acids whose side chains are not completely fixed.
A further reduction in the number of variables would be possible if one could predict from the amino acid sequence which parts of the molecule are in the form of alpha helixes. The few proteins whose structures have been completely determined provide some indication of which amino acids are likely to be found in helical regions, but not enough is yet known to make such predictions with any assurance.
Because protein chains are formed by linking molecules that belong to a single class (the amino acids), the linkage process can be expressed mathematically in a form that is particularly suited for a high-speed digital computer. We have written computer programs that calculate the coordinates of each atom of the protein, using as input variables only those angles in which rotational changes can occur; all other angles and bond lengths are entered as rigid constraints in the program. The method of calculation involves the use of a local coordinate system for the atoms in each amino acid unit and a fixed overall coordinate system into which all local coordinates are transformed.
The transformation that relates the local coordinate systems to the fixed coordinate system is recalculated by the computer program each time a new atom is added to the linear peptide backbone. Eaeh chemical bond is treated as a translation and a rotation of this transformation. The process requires a substantial amount of calculation, but each time the backbone reaches the central atom of a new amino acid the relative positions of the sidegroup atoms can be taken from the computer memory where this information is stored for each of the 20 varieties of amino acid. It is then a simple mutter to translate and rotate the side-group atoms from their local coordinate system into the fixed-coordinate system of th entire molecule.
The new value of the transformation at each step along the backbone is detetmined by the fixed rotation angles and translation distances that are built into the computer program and by the variable angles that must somehow be determined during the running of the program. The principal problem, therefore, is precisely how to provide correct values for the variable angles. A number of investigators are working on this problem in different ways. Before discussing any of these approaches I should emphasize the magnitude of the problem that remains even after one has gone as far as possible in using chemical constraints to reduce the number of variables from several thousand to a few hundred. Because each bond angle must be specified with an accuracy of a few degrees, the number of possible configurations that can result when each angle is varied by a small amount becomes astronomical even for a small protein. Moreover, these small rotations can produce a large effect on the total energy of the structure [see Figure 10].
One way to understand the difficulty of finding a configuration of minimum energy is to imagine the problem facing a man lost in a mountainous wilderness in a dense fog. He may know that within a few miles there is an inhabited river valley leading into a calm lake. He may also know that the lake is at the lowest point in the area, but let us assume that he- can only determine his own elevation and the slope of the ground in his immediate vicinity. He can walk down whatever hill he is standing on, but this would probably trap him nt the hottom of a small valley far from the lake he seeks. ln finding a configuration of minimum energy the comparable situation would be getting trapped in a metastable state far from a real energy minimum. Our lost man has only two dimensions to worry about, north-south and east-west; the corresponding problem in a molecule involves several hundred dimensions.
Our approach to this problem has assumed that even sophisticated techniques for energy minimization will not, at least at present, be sufficient to determine the structure of a protein from its amino acid sequence in a fully automatic fashion. We therefore decided to develop programs that would make use of a man-computer combination to do a kind of model-building that neither a man nor a computer could accomplish alone. This approach implies that one must be able to obtain information from the computer and introduce changes in the way the program is running in a span of time that is appropriate to human operation. This in turn suggests that the output or the computer must be presented not in numbers but in visual form.
I first became aware of the possibilities of using visual output from a computer in a conversation with Robert M. Fano, the director of Project MAC at the Massachusetts Institute of Technology. (MAC stands for multiple-access computer.) It soon became clear that the new types or visual display that had been developed would permit direct interaction of the investigator and a molecular model that was being constructed by the computer. All our subsequent work on this problem has made use of the large computer of Project MAC, which operates on the basis of "time-sharing," or access by many users. The system, developed by Fernando J. Corbato, allows as many as 30 people to have programs running on the computer at the same time. For all practical purposes it seems to each of them that he is alone on the system. A user can have any of his data in the high-speed memory printed out on his own typewriter, and he can make whatever changes he wants in this stored data. What is more important from our point of view is the ability to make changes in the commands that control the sequential flow of the program itself.
It is true, of course, that one of 30 users has access to a computer that, when it is fully occupied, has only a thirtieth of the speed of the normal machine, but for many problems this is enough to keep the man side of the man-machine combination quite well occupied. The program that acts to supervise the time-sharing system is organized in such a way that no user can interfere with the system or with any other user, and the computer is no allowed to stand idle if the man takes time out to think.
In working with molecular models we are interested in being able to obtain data quickly in order to evaluate the effect of changing the input variables of the program. For any particular molecular configuration the computer can readily supply the positions (in the three-dimensional coordinatcs x, y and z) of all the atoms. The important question is: What can be done with 5,000 to 10,000 numbers corresponding to the position of every atom in even a small protein? Obviously if we could formulate specific questions concerning energy, bond angles and lengths, overall shape, density and so on, the compulter could calculate the answer and there would be no need for a man ever to look ut the numerical values of the coordinates. We realized that our best hope of gaining insight into unexpected sstructural relations - relations that had not been anticipated - lay in getting th computer to present a thee-dimensional picture of the molecule.
Although computer-controlled oscilloscopes have been available for about 10 years, they have been used mainly to display numbers and letters. It is only recently that they have been used to produce the output of computer programs in grnphical form. The oscilloscope tube can of course present only a two-dimensional image. It is no great trick, however, to have the computer rotate the coordinates of the molecule before plotting their projection. If this is done, the brain of the human viewer readily constructs a three-dimensional image from the sequential display of two-dimensional images. Such sequential projectious seem to be just as useful to the brain as simultaneous stereoscopic projections viewed by two eyes. The effect of rotation obtained from the continously changing projection none the less has an inherent ambiguity. An observer cannot determine the direction of rotation from observation of the changing picture alone. In the Project MAC display system, designed by John Ward and Robert Stotz, the rate of rotation of the picture is controlled by the position of a globe on which the observer rests his hand; with a little practice the coupling between hand and brain becomes so familiar that any ambignuty in the picture can easily be resolved.
In evaluating the energy of a particular configuration of a protein molecule one must add up all the small interactions of atomic groups that contribute to this energy. These must include interactions not only between the different parts of the protein molecule but also between the parts of the protein molecule and the surrounding water molecules. 1f we are interested in altering the configuration in the direction of lower energy, we must be able to calculate the derivative of each of the energy terms - that is, the direction in which the energy curves slope - as changes are made in each or the rotation angles. Accordingly we must calculate a very large number of interatomic distances and the derivative of these distances with respect to the allowed rotation angle.
The derivatives can be calculated, however, without going through the extended transformation calculations needed to generate the coordinates themselves. The rotation around any chemical bond will cause one part of the molecule to revolve with respect to another. If both members of a pair of atoms are on the same side of the bond being altered, the rotation will not give rise to a change in their distance. If the two atoms are on opposite sides or the rotating bond, one of the atoms will move in a circle around lhe axis of rotation while the other remains stationary. By analyzing the geometry of this motion we can simplify the derivative calculations to the point where they require very little computation.
Even though each of these derivative calculations can be done in a few hundred microseconds of computer time, there would still be an excessive umount of calculation if it were done for all possible pairs of atoms within the molecule. Fortunately the interactions with which we are concerned are short-range ones, and most of the pairs of atoms are too far apart to contribute appreciably to the overall energy. In order to select out all pairs of atoms that are close to each other, we have developed a procedure called cube-testing. All the space in the region of the molecule is divided into cubes of some predetermined size, let us say five angstroms on an edge (two or three times the typical bond length) and each atom is assigned to a cube. To consider the interactions involving any given atom one need only determine the distance between this atom and all others in the same cube and in the 26 surrounding ones [see Figure 11]. Although the procedure is still time-consuming, it is much faster than having to do calculations for all possible pairs of atoms in the molecule.
In addition to enabling us to screen the data for close pairs, the cubing procedure provides information about which groups in the protein molecule can interact with the surrounding water molecules. In order to enumerate such interactions, we first deflne the "insideness" and "outsideness" of the molecule, outsideness meaning that a particular atom or group of atoms is accessible to the surrounding water and insideness that it is not. If we examine thee cubing pattern for a particular molecule, an atom on the outside would be in a cube that is surrounded on one side by filled cubes and on the other hy empty ones. In a similar way we can detect holes in the midst of the structure by looking for empty cubes that are surrounded on all sides by filled ones.
By suitable use of derivative caleulation and cubing we can alter any configuration of the molecule in the direction of lower energy. This procedure, however, would almost certainly lead to a structure that is trapped in one of th local minima - one of the highcr valleys of our wilderness analogy - and may not even be close to the true minimum-energy configuration we are looking for. For a real molecule floating in solution, the local minima would not represent traps because the normal thermal vibration of the molecule and its parts supplies euongh energy to move Lhe structure out of any valley that is not a trune minimum.
Although there are various ways in which one can use random elements in a computer calculation to simulate thermal vibration, it is our experience that an investigator who is looking at the molecule can frequently understand the reason for the local minimum and by making a small alteration in the structure can return the program to its downhill path. Such alterations can b accomplished in the computer by changing the program in such a way as to introduce pseudo-energy terms that have the effect of pulling on parts of the structure. A few simple subprograms that introduce the appropriate pseudo-energies enable us to do the same kind of pulling and pushing in the computer that we can do with our hands while building actual models.
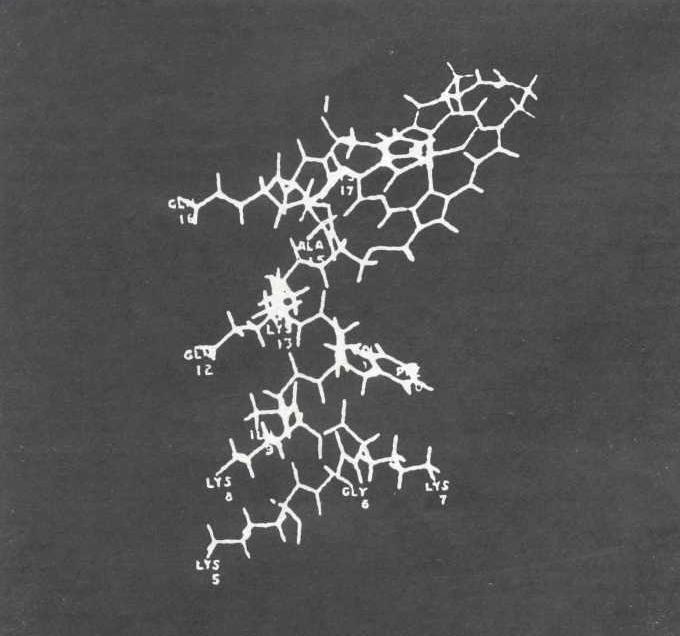
Pulling a structure by means of these pseudo-energy terms is also useful for building a model that observes all the chemical constraints and at the same time has its atoms as close as possible to the positions indicated by X-ray diffraction studies. In this case the pseudo-energies can be regarded a springs pulling designated atoms to their experimentally determined positions. Such calculations have been carried out by Martin Zwick, a graduate student at M.I.T., in order to make a model of myoglobin fit the configuration determined from X-ray data hy John C. Kendrew and Herman Watson at the Laboratory of Molecular Biology in Cambridge. For this type of problem a helpful procedure has been developed by William Davidon of Haverford College. In his program one starts by "walking" in the direction of the steepest slope, but with each successive step one builds up information as to how the slope of the hillside changes.
Once we had produced a computer model of myoglobin, we could ask questions concerning the relative importance of short-range forces acting between various parts of the molecule. There are, for example, Van der Waals forces: electrostatic attractions due to the electric dipoles that all atoms induce in one another when they are close together. There are also electrostatic interactions that arise from the permanent electric dipoles associated with the peptide bonds. The permanent dipole attractions turned out to be larger than we had expected. It is thus possible that the electrostatic: interaction between different regions of a protein may play a substantial role in stabilizing its structure. This results in part from the fact that the electric dipoles in an alpha helix are added to one another along the direction of the axis. For this reason two helixes that wind in the same direction will repel each other and two that wind in opposite directions will attract each other. In myoglobin the overall effect is a substantial attractive force. This calculation requires some form of model-building, because the electrically charged regions are associated with the C-0 and N-H groups along the backbone, and the hydrogcn atom is not detected in the X-ray analysis.
Although the electrostatic interaction energy is of the same order of magnitude as that required to denature a protein, it is probably not the dominant energy for the folding or a protein molecule. The primary source of energy for this purpose probably comes from the interaction of the amino acid side chains and the surrounding water; the electrostatic interactions may do no more than modify the basic structure.
We still have much to learn about the magnitude of the various energy terms involved in holding a protein molecule together. Meanwhile we have been trying to develop our computer technique, using the knowledge we have. Is this knowledge enough to enable us to find the lowest energy state of a protein molecule and to predict its structure in advance of its determination by X-ray analysis?
The answer to this question probably depends on how well we understand what really happens when a protein molecule in a cell folds itself up. Our work has been based on the hypothesis that the folding starts independently in several regions of the protein and that the first structural development is th formation of a number of segments of alpha helix. Our assumption is that these segments then interact with one another to form the final molecular structure. In this method of analyzing the problem the units that have to be handled independently are the helical regions rather than the indiviclual amino acids. Thus the number of independent variables is greatly reduced. The success of the proceedure depends, however, on the assumption that we can deduce from the amino acid sequence alone which regions are likely to be helical. It is not necessary, however, that we guess the helical regions correctly the first time we can see what happens when helical regions are placed in many different parts of the molecule. Several other groups working on this problem are following the hypothesis that the folding proceeds only from the amino end of the protein. Until one of these approaches succeeds in predicting the structure of a protein and having the prediction confirmed by X-ray analysis, we can only consider the different hypotheses as more or less plausible working guides in studying the problem.
It is still too early to evaluate the usefulness of the man-computer combination in solving real problems of molecular biology. It does seem likely, however, that only with this combination can the investigator use his "chemical insight" in an effective way. We already know that we can use the computer to build and display models of large molecules and that this procedure can be very useful in helping us to understand how such molecules function. But it may still be a few years before we have learned just how useful it is for the investigator to be able to interact with the computer while the molecular model is being constructed.
CLETC ACID (DNA is modeled by a program nd.rew A. MacEwan of Lhe hildren's Cancer Research Foundation and Harvard Medical chool. This DNA sequence left to right, top to bottom) begins with a single nucleotide: a pentagonal sugar plus a phosphate group and a base. A second nucleotide is added, and then more to make a helical chain that joins with a second chain to form the characteristic double helix. Then the helix is rotated in space.









